The Research Group Dynamics of Social Behavior (DYNOSOB 🦖) was created only a few years ago. In the earlier years of the group it was important to us that we set some “rules” and create a somewhat of a common workflow. The rules are regarding the way we do research and publish scientific results, and the main reason we wanted to set these rules was reproducibility.
Reproducibility is the trademark of good science, and on that note the use of tools, such as version control and hosting platforms for version control, are becoming vastly used.
In this blog post we would like to outline the “rules” and workflow we established for our research group. We decided to use the version control system git and the hosting platform GitHub. git was the obvious choice since one of our group members is very familiar with the tool, and we decided to use GitHub over GitLab because it was easier for us to collaborate with external collaborators via GitHub.
Introduction to the tools. Any new member that joins our group has to complete a workshop on “Introduction to Research Software Development”. This workshop is intended for complete beginners and it covers:
- Introduction to the command line
- Introduction to Python
- Best practices for writing software (with Python examples)
- Version control with git
- Using GitLab and GitHub
A new member can either cover the material on their own time, or attend one of the annual workshops offered by Nikoleta (group member) and Carsten Fortmann-Grote (head of the scientific computing unit at the Max Planck Institute for Evolutionary Biology).
After a new member has completed the workshop they are asked to create a personal profile and add it to the group’s website. The source code for the website is hosted on GitHub (repository) and in order to add a profile one needs to use git to clone the project, commit their changes and open a pull request. We use this as an exercise to give new members a more “realistic” experience with the git and GitHub.
Housekeeping. As you might have noticed the website’s repository is part of the organisation DYNOSOBs which is the organisation used to host any group related repositories.
The organisation includes the following repositories (as of today):
- DYNOSOB-talks: contains presentation slides and posters of the group.
- DYNOSOB-bibliography: contains a shared bibliography.
- DYNOSOB-tutorials: a collection of examples and tutorials of techniques that we have put together.
The organisation is also used to host a copy of project related repositories. This brings us to the next part.
Conducting science & publishing results. The members of our group are expected to use git to manage their research projects, and moreover each research project is expected to have a corresponding GitHub repository.
The learning curve for git can be quite steep in the beginning. For this reason we have put together a flow chart with commands that we find useful (please let us know if we can improve or if we forgot something):
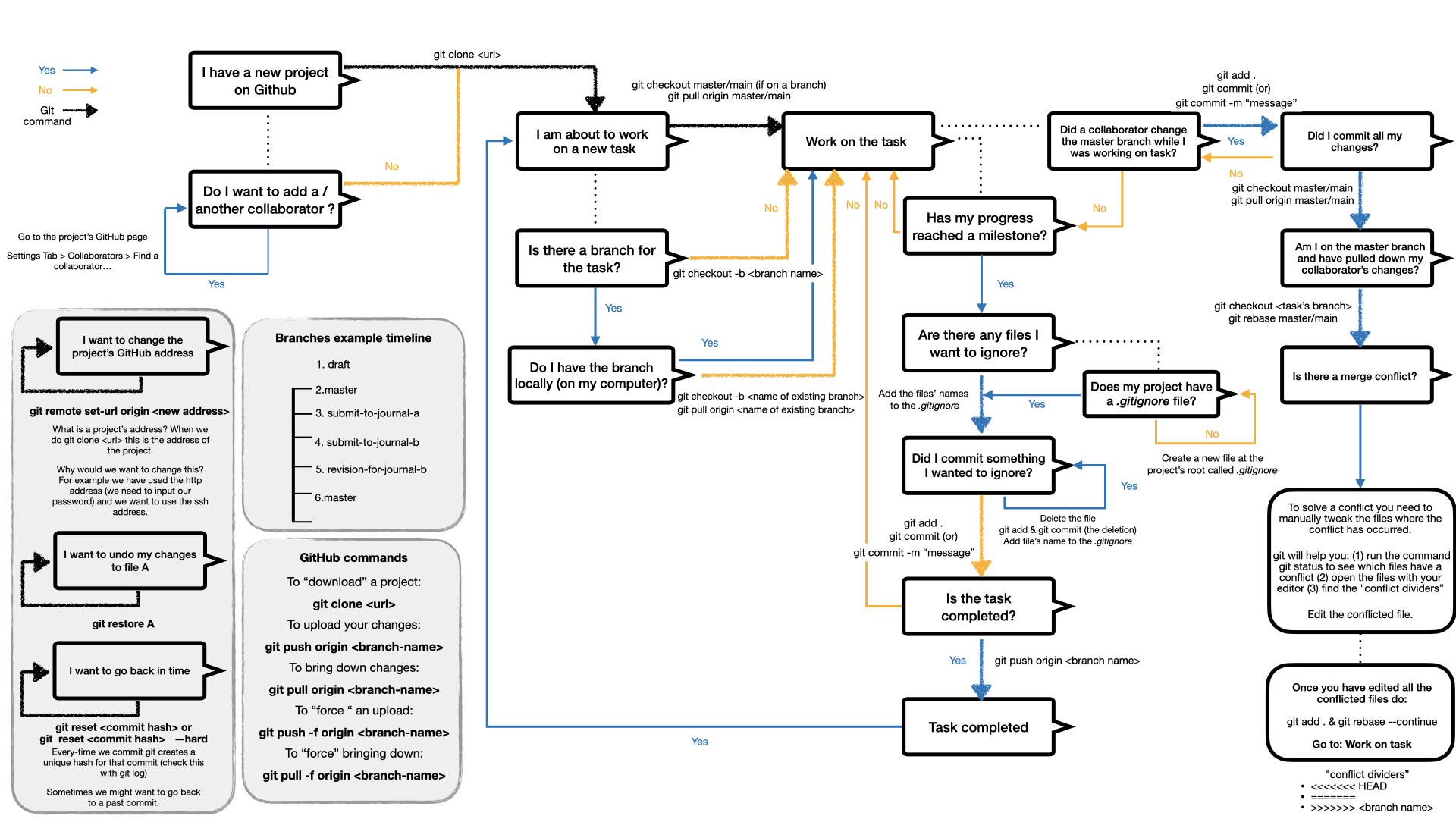
A research project’s GitHub repository is expected to contain all the code that was used to produce the scientific results, and the files necessary to compile the manuscript (we write our papers in LaTex). The repositories are usually kept private while we work on projects and once a paper has been published we make them public. Following a publication the members are asked to create a copy of that repo under the organisation DYNOSOBs. That way even if the member leaves or deletes their GitHub profile the source material can still be found.
Note that we do not use GitHub to host the data produced by our simulations, or from our experiments. For data we use websites such as Zenodo and Open Science Framework.