I am a group leader at the Turing Center for Living Systems (CENTURI), an interdisciplinary center attached to Aix-Marseille Université, in the South of France, by the sea. My group is based at the Laboratoire de Chimie Bactérienne, at the Joseph Aiguier CNRS Campus. My research uses statistical physics and mathematical modeling to study biological systems. I am particularly interested in microbial communities that live in association with hosts, a.k.a. microbiomes, and aim at addressing a wide range of related questions, like:
- How do hosts and microbes co-evolve?
- How is the microbiota diversity maintained in the host?
- What are the control mechanisms that the host can exert on its microbiota at minimal costs?
- How can within-host microbial dynamics impact an infectious spread at the scale of the host population?
- Can microbiota modeling provide valuable insights for clinical applications or to public health decision makers?
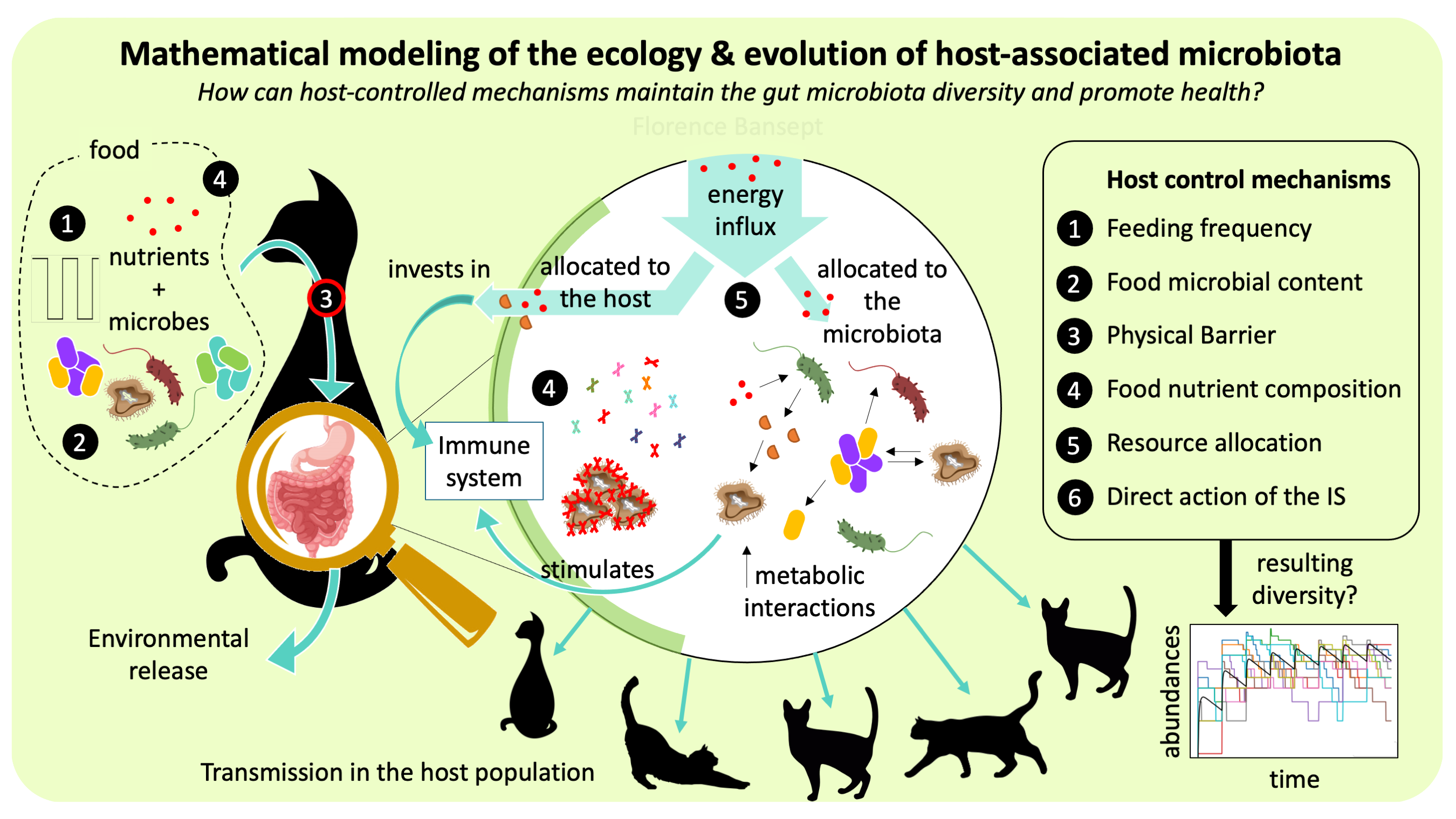
To study these questions, I use a combination of analytical and numerical techniques with stochastic simulations and collaborate and frequently exchange with experimentalists.
Research on microbiome dynamics presents specific challenges. As meta-omics enable the gathering of more and more observational data, mechanistic modeling – that is, modeling that proposes causal mechanisms to explain observed data – remains crucially needed. One key challenge that requires particular efforts and that I find particularly exciting is the inference of dynamical parameters from indirect snapshot data – a problem similar to the one of a detective having to reconstitute what has happened from the clues on the crime scene only.
List of Publications
Peer-reviewed publications
- N. Obeng, A. Czerwinski, D. Schütz, J. Michels, J. Leipert, F. Bansept, M. J. García García, T. Schultheiß, M. Kemlein, J. Fuß, A. Tholey, A. Traulsen, H. Sondermann & H. Schulenburg, Bacterial c-di-GMP has a key role in establishing host–microbe symbiosis, Nature Microbiology, 2023
- D. Hoces, G. Greter, M. Arnoldini, M.L. Stäubli, C. Moresi, A. Sintsova, S. Berent, I. Kolinko, F. Bansept, A. Woller, J. Häfliger, E. Martens, W-D. Hardt, S. Sunagawa, C. Loverdo, E. Slack, Fitness advantage of Bacteroides thetaiotaomicron capsular polysaccharide in the mouse gut depends on the resident microbiota, eLife, 2023
- R. Zapién-Campos, F. Bansept, M. Sieber, and A. Traulsen, On the effect of inheritance of microbes in commensal microbiomes, BMC Ecology and Evolution, 2022
- F. Bansept, N. Obeng, H. Schulenburg, and A. Traulsen, Modeling host-associating microbes under selection, The ISME Journal, 2021
- N. Obeng, F. Bansept, M. Sieber, A. Traulsen, H. Schulenburg, Evolution of microbiota-host associations: the microbe’s perspective, Trends in Microbiology, 2021
- F. Bansept, L. Marrec, A-F. Bitbol and C. Loverdo, Antibody-mediated cross-linking of gut bacteria hin- ders the spread of antibiotic resistance, Evolution 73-6: 1077–1088, 2019 ( arXiv:1903.05723v1)
- F. Bansept, K. Moor, M. Diard, W-D Hardt, E. Slack and C. Loverdo, Enchained growth and cluster dislocation: a possible mechanism for microbiota homeostasis, PLoS Comput Biol 15(5):e1006986, 2019
- K. Moor, M. Diard, M. E. Sellin, B. Felmy, S. Y. Wotzka, A. Toska, E. Bakkeren, M. Arnoldini, F. Bansept, A. Dal Co, T. Völler, A. Minola, B. Fernandez-Rodriguez, G. Agatic, S. Barbieri, L. Piccoli, C. Casiraghi, D. Corti, A. Lanzavecchia, R. R. Regoes, C. Loverdo, R. Stocker, D. R. Brumley, WD Hardt, E. Slack, High-avidity IgA protects the intestine by enchaining growing bacteria, Nature, 544(7651):498-502, 2017
Preprints and manuscripts in preparation
- A. Galeev, F. Bansept, N. Benny, V. Chaves Vargas, A. Habich, A. Traulsen, D. Unterweger, Type VI secretion systems of Pseudomonas aeruginosa modulate infection dynamics and immune responses in Galleria mellonella larvae, in revision
- R. Zapién-Campos, F. Bansept, A. Traulsen, Inferring interactions from microbiome data, submitted (preprint on bioRxiv)
Thesis
-
My PhD thesis entitled "Biophysical modeling of bacterial population dynamics
& the immune response in the gut" is available here.
About me

Before I fell into Biology, I studied physics and a little bit of musicology at École Normale Supérieure in Paris.
During my PhD with Claude Loverdo, I worked on several aspects of bacterial population dynamics and the interactions with the immune system in the gut. We collaborated with the Slack lab where Salmonella infections in mice are studied. We contributed to evidence “enchained growth”, the mechanism by which a specific antibody – Immunoglobulin A (IgA), produced in large quantities following vaccination – enchains dividing daughter-bacteria in clonal clusters and protects the mice from falling sick. We showed that enchained growth could potentially allow the immune system to maintain homeostasis of the microbiota composition. At the scale of the host population, it could hinder the spread of antimicrobial resistance, thus advocating for sustained efforts in the development of IgA-inducing vaccines.
After my PhD, I expatriated and went on a postdoc working in the group of Arne Traulsen at the Max Planck Institute for Evolutionary Biology in Plön, Germany. My research took a more evolutionary turn and I developed models studying the selection on the life-history traits of host-associating microbes, by importing methods from demography to the field of microbiota research. I focused on the specificity of biphasic life cycles - where microbes transition from the environment to the host and back - and assessed the cases in which the role of migration prevails over the one of replication for microbial fitness. From mice I went to worms, stickleback fishes, moth larvae, drosophila and many more, having established fruitfull collaborations with multiple members of the Collaborative Research Center Origin and Function of Metaorganisms, in Kiel, Germany, where I was also affiliated for four years.

In March 2023, I am starting my own group at the Turing Center for Living Systems, where I will continue investigating the mathematical rules underlying microbiota dynamics. Look out for positions to be announced soon!

I also enjoy teaching, and I have taken part in several outreach projects. In particular, I have been involved in the development of an educational website for the general public related to the COVID-19 pandemic. One of the activities we developed for this website has been funded by the Robert Bosch Stiftung to be produced as a proper board game, distributed in over 10 000 copies over more than 5 countries! It is available for free and in several languages here.
You can download my complete CV here.

In my spare time, I am also a keen amateur musician. My first instrument is the piano, but I also study opera singing at Marseille Conservatoire. At the moment, I am involved in two vocal ensembles: the Choeur Minuscule, of which I am one of the artistic co-directors, and the semi-professional feminine vocal ensemble Hymnis. In the past, I occasionally contributed musical critiques to the French website ComposHer, which promotes women and particularly women composers in classical music.
