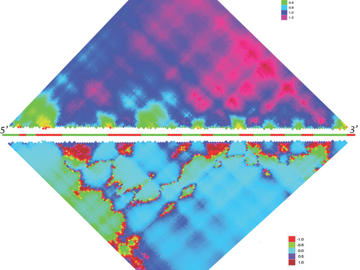
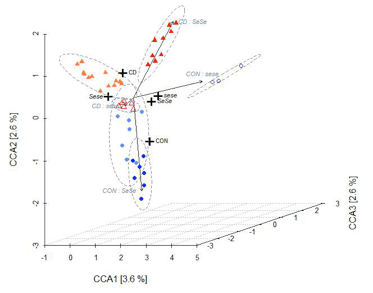
Our overall aim is to understand the interplay between mammalian hosts and their associated microbial communities from an evolutionary perspective in order to gain novel insight into human health and disease. We use a combination of evolutionary-, functional- and metagenomic analysis to understand origins and consequences of genetic variation of both the host and its microbes. Our work includes both the house mouse as a model organism as well as human studies based at the University Clinic Schleswig-Holstein (UKSH).
Evolutionary and metagenomic analysis of candidate genes involved in homeostasis of the gastrointestinal tract
We are investigating the evolution of disease-associated genes and their influence on the intestinal microbiota in both humans and mice. Of particular interest are blood-group related glycosyltransferases, as they are highly expressed in the GI tract, are frequent targets of selection, and are often associated with trade-offs in relation to resistance to pathogens and susceptibility to disease. Current work focuses on the glycosyltransferases FUT2 in humans an B4galnt2 in house mice, both of which are targets of balancing selection and associated disease. This work is currently funded by the German Science Foundation (DFG) Priority Program 1656, "Intestinal Microbiota- A Microbial Ecosystem at the Edge between Immune Homeostasis and Inflammation."
Click for Representative Publications
Rausch, P., Steck, N., Suwandi, A., Seidel, J.A., Künzel, S., Bhullar, K., Basic, M., Bleich, A., Johnsen, J.M., Vallance, B.A., Baines, J.F. and Grassl, G.A. (2015), “Expression of the Blood-Group-Related Gene B4galnt2 Alters Susceptibility to Salmonella Infection”, PLoS Pathogens, Vol. 11 No. 7 p. e1005008. doi: 10.1371/journal.ppat.1005008.
Staubach, F., Künzel, S., Baines, A.C., Yee, A., McGee, B.M., Bäckhed, F., Baines, J.F. and Johnsen, J.M. (2012), “Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice”, The ISME Journal, Vol. 6 No. 7, pp. 1345–1355. doi: 10.1038/ismej.2011.204.
Rausch, P., Rehman, A., Künzel, S., Häsler, R., Ott, S.J., Schreiber, S., Rosenstiel, P., Franke, A. and Baines, J.F. (2011), “Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 108 No. 47, pp. 19030–19035. doi:10.1073/pnas.1106408108.
Linnenbrink, M., Johnsen, J.M., Montero, I., Brzezinski, C.R., Harr, B. and Baines, J.F. (2011), “Long-term balancing selection at the blood group-related gene B4galnt2 in the genus Mus (Rodentia; Muridae)”, Molecular Biology and Evolution, Vol. 28 No. 11, pp. 2999–3003. doi:10.1093/molbev/msr150.
Staubach, F., Künzel, S., Baines, A.C., Yee, A., McGee, B.M., Bäckhed, F., Baines, J.F. and Johnsen, J.M. (2012), “Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice”, The ISME Journal, Vol. 6 No. 7, pp. 1345–1355. doi: 10.1038/ismej.2011.204.
Rausch, P., Rehman, A., Künzel, S., Häsler, R., Ott, S.J., Schreiber, S., Rosenstiel, P., Franke, A. and Baines, J.F. (2011), “Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype”, Proceedings of the National Academy of Sciences of the United States of America, Vol. 108 No. 47, pp. 19030–19035. doi:10.1073/pnas.1106408108.
Linnenbrink, M., Johnsen, J.M., Montero, I., Brzezinski, C.R., Harr, B. and Baines, J.F. (2011), “Long-term balancing selection at the blood group-related gene B4galnt2 in the genus Mus (Rodentia; Muridae)”, Molecular Biology and Evolution, Vol. 28 No. 11, pp. 2999–3003. doi:10.1093/molbev/msr150.
Host-microbiota coevolution in the mammalian intestine
This project aims to understand the forces that shape variation in host-associated bacterial communities within and between host species, which is key to understand the evolution and maintenance of metaorganisms. The house mouse subspecies Mus musculus domesticus and M. m. musculus share a common ancestor about 0.5 MYA, yet remain only partially reproductively isolated. This enables the divergence in host-microbiota interactions that have occurred since their common ancestor to be studied using genetic approaches.To identify and characterize host loci contributing to differences in the microbiota between host species, we perform quantitative trait locus (QTL) mapping in inbred strains as well as association mapping in wild-derived hybrids. In parallel, we are further investigating candidate bacterial traits using a combination of shotgun metagenomic sequencing and single-cell/ isolate analysis to characterize them at the functional level and identify potential signatures of adaptation to their hosts. This project was awarded funding through the recently established DFG Collaborative Research Center (CRC) 1181, "Origin and Function of Metaorganisms." (press release)
Click for Representative Publications
Wang, J., Kalyan, S., Steck, N., Turner, L.M., Harr, B., Künzel, S., Vallier, M., Häsler, R., Franke, A., Oberg, H.H., Ibrahim, S.M., Grassl, G.A., Kabelitz, D. and Baines, J.F. (2015), “Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome”, Nature Communications, Vol. 6, p. 6440. doi:10.1038/ncomms7440.
Linnenbrink, M., Wang, J., Hardouin, E.A., Künzel, S., Metzler, D. and Baines, J.F. (2013), “The role of biogeography in shaping diversity of the intestinal microbiota in house mice”, Molecular Ecology, Vol. 22 No. 7, pp. 1904–1916. doi:10.1111/mec.12206.
Linnenbrink, M., Wang, J., Hardouin, E.A., Künzel, S., Metzler, D. and Baines, J.F. (2013), “The role of biogeography in shaping diversity of the intestinal microbiota in house mice”, Molecular Ecology, Vol. 22 No. 7, pp. 1904–1916. doi:10.1111/mec.12206.
The role of host gene x microbiota interactions in susceptibility to autoimmune skin blistering
In collaboration with the Lübeck Institute for Experimental Dermatology, we are investigating the role of skin microbiota in the origin and severity of Pemphigoid diseases, which are characterized by autoimmunity to structural (collagen) proteins located at the dermal and epidermal junction. First, using an advanced, four-way inter-cross between resistant and susceptible mouse stains, we are performing quantitative trait locus (QTL) mapping to identify host genes influencing disease and/or interactions with skin microbes. This and other experimental mouse models reveal a probiotic effect of members of the skin microbiota, for which cultivation and characterization is underway. This work is supported by the DFG Research Training Group 1743, "Genes, Environment, and Inflammation." Second, we are now investigating these phenomena and their therapeutic potential in human cohorts as part of the Clinical Research Unit 303, "Pemphigoid Diseases: Molecular Pathways and their Therapeutic Potential, funded by the DFG.
Click for Representative Publications
Ellebrecht, C.T., Srinivas, G., Bieber, K., Banczyk, D., Kalies, K., Künzel, S., Hammers, C.M., Baines, J.F., Zillikens, D., Ludwig, R.J. and Westermann, J. (2015), “Skin microbiota-associated inflammation precedes autoantibody induced tissue damage in experimental epidermolysis bullosa acquisita”, Journal of Autoimmunity, doi:10.1016/j.jaut.2015.08.007.
Wang, J., Kuenzel, S. and Baines, J.F. (2014), “Draft Genome Sequences of 11 Staphylococcus epidermidis Strains Isolated from Wild Mouse Species”, Genome Announcements, Vol. 2 No. 1, doi:10.1128/genomeA.01148-13.
Srinivas, G., Möller, S., Wang, J., Künzel, S., Zillikens, D., Baines, J.F. and Ibrahim, S.M. (2013), “Genome-wide mapping of gene–microbiota interactions in susceptibility to autoimmune skin blistering”, Nature Communications, Vol. 4, doi:10.1038/ncomms3462.
Wang, J., Kuenzel, S. and Baines, J.F. (2014), “Draft Genome Sequences of 11 Staphylococcus epidermidis Strains Isolated from Wild Mouse Species”, Genome Announcements, Vol. 2 No. 1, doi:10.1128/genomeA.01148-13.
Srinivas, G., Möller, S., Wang, J., Künzel, S., Zillikens, D., Baines, J.F. and Ibrahim, S.M. (2013), “Genome-wide mapping of gene–microbiota interactions in susceptibility to autoimmune skin blistering”, Nature Communications, Vol. 4, doi:10.1038/ncomms3462.
Last updated May 2023
© John F. Baines 2023
© John F. Baines 2023
Create a free web site with Weebly