
Position: Project Leader
Office: 121
Office phone: + 49 4522 763-237
E-mail: pboynton[a]evolbio.mpg.de
Twitter: @pjboynton
Research interests
I use yeasts to understand microbial evolution and ecology in nature. Yeasts are diverse single-celled fungi, and can be found in almost every imaginable habitat. I currently work with the most famous group of yeasts, Saccharomyces yeasts.
The genus Saccharomyces includes domesticated baker’s yeast, S. cerevisiae, which is found in bread, beer, wine, and a variety of other fermented foods and vegetables. Saccharomyces also includes several wild yeast species that live in forests. I study S. paradoxus, the closest wild relative of S. cerevisiae, which lives on soil and leaf litter in local German forests (Boynton et al. 2019) (Figure 1).
I am using S. paradoxus to understand past, present, and future microbial evolution. S. paradoxus has been evolving, and continues to evolve, in complicated and changing natural environments with diverse chemicals, other organisms, and environmental stresses. I would like to understand how S. paradoxus has adapted to its environment and how the environment continues to change S. paradoxus. A lot of what we know about microbial evolution comes from laboratory studies of model organisms, including S. cerevisiae. This laboratory experimental evolution has taught us how microbes can evolve, but we have to look to natural populations to understand how microbes have evolved. My research combines both strategies to understand how S. paradoxus is evolving in nature.

Figure 1: Saccharomyces paradoxus leaf litter habitat near the Max-Planck Institute in Plön. S. paradoxus is found in soil and on decomposing leaves beneath oak trees. Photo by Duncan Greig
Current projects
Temporal adaptation in S. paradoxus
S. paradoxus is subject to seasonal changes in temperature and nutrients in its natural environment. I would like to understand how the fitness of an individual yeast cell responds to these environmental changes and interactions. In particular, I am looking for seasonally locally adapted genotypes that have high fitness in a single season, and phenotypically plastic genotypes that have high fitness in several environments.
Fitness consequences of inbreeding in nature
S. paradoxus can mate with close relatives (such as siblings) or unrelated individuals during its sexual life cycle, but it appears to preferentially mate with siblings. I am supervising the Master’s thesis project of Onur Erk Kavlak, in collaboration with Michael Sieber of the Evolutionary Theory department, to ask why S. paradoxus inbreeds so frequently. We are combining a field experiment with evolutionary theory to understand what kinds of natural environmental conditions select for inbreeding.
Naturally-occurring killer yeasts
“Killer” yeasts produce extracellular killer toxins, which kill nearby sensitive yeasts (Boynton 2019) (Figure 2). These toxins can help a killer yeast by removing competitors from the environment, at least in the lab. But how do these killer yeasts interact with other yeasts in nature? I recently supervised the Master’s thesis project of Rahul Unni to understand the effects of toxins on toxin producers and other yeasts in the environment.
Figure 2: Example of killer yeasts. Drops of two different S. paradoxus yeasts were put onto petri dish covered with a third yeast. The drop on the left produces a toxin that kills nearby cells, while the drop on the right does not.
S. paradoxus natural history
Saccharomyces yeasts are famous for their love of sugars, but natural S. paradoxus soil and leaf litter environments have little available sugar. I am supervising the Master’s thesis of Antonia Reinhold, in collaboration with Tobias Demetrowitsch of the Kiel University Institute of Human Nutrition and Food Science to understand the chemical environments in which S. paradoxus lives and grows.
Other projects and interests
The community ecology of dominant species
Yeasts live in diverse communities, surrounded by other microbes. In many communities, a single yeast species will become dominant (i.e., the species will be more common than any other yeast in the community). For example, the yeast Candida pseudoglaebosa is dominant in pitchers of the carnivorous plant Sarracenia purpurea (Figure 3) because it arrives in pitchers early in pitcher development, before other yeasts (Boynton et al. 2019). This early arrival allows C. pseudoglaebosa to be the most common yeast in a group of pitchers long after it arrives, even after better competitors arrive in the pitchers. In contrast S. cerevisiae, the famous domesticated yeast, becomes dominant in wine fermentations because it is a very good competitor: it outcompetes other yeasts in the fermentation. High species diversity can prevent S. cerevisiae from dominating. When there are too many other species, S. cerevisiae does not become dominant, and grape juice does not ferment to wine (Boynton & Greig 2016).

Figure 3: Sarracenia purpurea produces carnivorous pitchers that fill with rainwater and house diverse microbial communities. The yeast Candida pseudoglaebosa is the dominant fungus in these microbial communities
Microbial local adaptation
Local adaptation has evolved when organisms from one environment are fitter in that environment than organisms from elsewhere. While local adaptation evolves quickly in laboratory evolution experiments, there are few examples of local adaptation among free-living microbes (Kraemer & Boynton 2017). My colleagues and I designed special chambers in which S. paradoxus from different environments could compete in leaf litter, and we used the chambers to look for evidence of local adaptation (Boynton et al. 2016). While there was no evidence for S. paradoxus local adaptation, we did find fitness differences among individuals with different copies of a protein used for bringing sugars into cells.
Yeast interspecies hybridization
Different yeast species occasionally mate and have hybrid offspring. Hybrid Saccharomyces are frequently found in domesticated environments (Figure 4) (Boynton & Greig 2014), even though very few hybrid matings produce surviving offspring. My colleagues and I, including Thijs Janzen from the Evolutionary Theory Department, designed a mathematical model to explain how mistakes during meiosis (the cell division leading to cells that can mate with one another) can prevent hybrid offspring from surviving (Boynton et al. 2018).
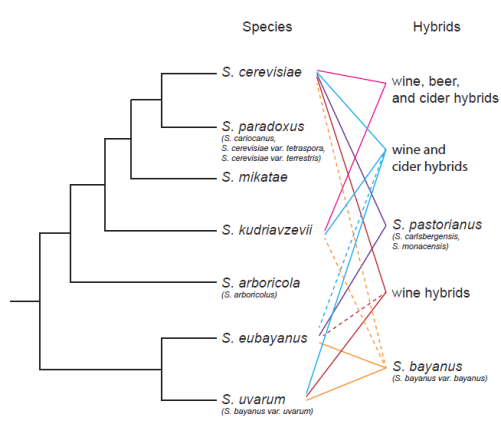
Figure 4: Saccharomyces cladogram showing phylogenetic relationships between S. paradoxus and its close relatives. Most Saccharomyces species are wild, but some S. cerevisiae and S. uvarum strains are domesticated and live in high-sugar environments. Many hybrid Saccharomyces are also found in domesticated environments. (Boynton & Greig 2014)
Short CV
2017-present: Project leader, MPI for Evolutionary Biology (Eva Stukenbrock’s lab)
2012-2017: Postdoctoral researcher, MPI for Evolutionary Biology (Duncan Greig’s lab)
2006-2012: Ph.D. Organismic and Evolutionary Biology, Harvard University, Cambridge, Massachusetts, USA (Anne Pringle’s lab)
2005-2006: Laboratory Technician, UC Berkeley (Thomas Bruns’s lab)
2003-2005: Natural Resource Management Agent, Peace Corps Togo
1998-2002: B.S. Genetics and Plant Biology, UC Berkeley (Thomas Bruns’s lab)
Publications
Forest Saccharomyces paradoxus are robust to seasonal biotic and abiotic changes. Boynton PJ, Wloch-Salamon D, Landermann D, Stukenbrock EH (preprint) BioRxiv doi:10.1101/2020.04.27.063289.
The ecology of killer yeasts: interference competition in natural habitats. Boynton PJ (2019) Yeast 36:473-485 doi: 10.1002/yea.3398.
Quantifying the efficiency and biases of forest Saccharomyces sampling strategies. Boynton PJ, Kowallik V, Landermann D, Stukenbrock EH (2019) Yeast 36:657-668 doi: 10.1002/yea.3435.
Superior dispersal ability can lead to persistent ecological dominance throughout succession. Boynton PJ, Peterson CN, Pringle A (2019) Applied and Environmental Microbiology doi: 10.1128/AEM.02421-18.
Modeling the contributions of chromosome segregation errors and aneuploidy to Saccharomyces hybrid sterility. Boynton PJ, Janzen T, Greig D (2018) Yeast 35:85-98, doi: 10.1002/yea.3282.
Evidence for microbial local adaptation in nature. Kraemer SA*, Boynton PJ* (2017) Molecular Ecology, 26:1860-1876, doi:10.1111/mec.13958. *joint first authors.
Measuring microbial fitness in a field reciprocal transplant experiment. Boynton PJ, Stelkens R, Kowallik V, Greig D (2017) Molecular Ecology Resources 17:370-380, doi: 10.1111/1755-0998.12562.
Species richness influences wine ecosystem function through a dominant species. Boynton PJ, Greig D (2016) Fungal Ecology 22:61-72, doi: 10.1016/j.funeco.2016.04.008.
Fungal diversity and ecosystem function data from wine fermentation vats and microcosms. Boynton PJ, Greig D (2016) Data in Brief 8:225-229, doi: 10.1016/j.dib.2016.05.038.
Age-related cellular changes in the long-lived bivalve A. islandica. Gruber H, Wessels W, Boynton P, Xu J, Wohlgemuth S, Leeuwenburgh C, Qi W, Austad SN, Schaible R, Philipp EE (2015) Age 37:90, doi: 10.1007/s11357-015-9831-8.
The ecology and evolution of non-domesticated Saccharomyces species. Boynton PJ, Greig D (2014) Yeast 31:449-462, doi: 10.1002/yea.3040.
Inoculum potential of Rhizopogon spores increases with time over the first 4 yr of a 99-yr spore burial experiment. Bruns TD, Peay KG, Boynton PJ, Grubisha LC, Hynson NA, Nguyen NH, Rosenstock NP (2009) New Phytologist 181:463-470, doi: 10.1111/j.1469-8137.2008.02652.x.